Research

We investigate the molecular mechanisms that allow organisms in general, and microbes in particular, to swiftly adapt to environmental changes and stress, either through genetic and epigenetic changes, or by adapting gene expression and exploiting biological noise and bet-hedging.
Our favorite model organism is Saccharomyces cerevisiae, a species of yeast used for centuries for baking, brewing and winemaking. We also explore evolution and adaptation in other model organisms from time to time, often in collaboration with other groups.

Systems biology
We focus on untangling genotype-phenotype interactions and use comparative genomics and experimental evolution to investigate how species adapt.
In doing so, we combine molecular biology, high-throughput technologies and computational methods.

Synthetic biology
We design and use synthetic biology tools to understand, improve and modify existing biological systems.
We engineer strains for the production of industrially relevant compounds, such as bioethanol, proteins, pharmaceuticals and green chemicals. Combining DNA synthesis and targeted mutagenesis, we can rapidly create and screen millions of potentially useful variants with our state-of-the-art high-throughput equipment (Singer RoToR, colony picker, Echo liquid handler, customized robotic platform for microbial handling and phenotyping).

Evolutionary genomics
We study DNA to gain insight into the patterns and processes of evolution.
We study the mechanisms underlying hyper-evolvable properties, which often lie at the border between genetics and epigenetics, including such phenomena as chromatin silencing, hyper-variable tandem repeats, transposable elements, telomeric recombination, and hysteresis in regulatory pathways.
Some properties of living organisms evolve and diverge at a much higher pace than other traits. Examples range from the molecular composition of the microbial cell surface to the skeletal morphology of dogs. Our lab studies the processes underlying such hyper-evolvable properties, using classic genetic techniques as well as genomics, bioinformatics and mathematical modeling.
Swift evolution can be seen as a mechanism for “short-term transgenerational memory”. In other words, parental cells can pass on certain information to their progeny, but only for a few generations. In this way, the “memory” stays flexible enough to deal with rapid (external) changes that might require cells to “ignore” the parental “advice”. We hypothesize that the genetic systems underlying hyper-evolvable traits often developed as a response to hyper-variable selective pressure. This implies that apart from standard genetics and biochemistry, we also incorporate evolution theory to help explain and understand our results.
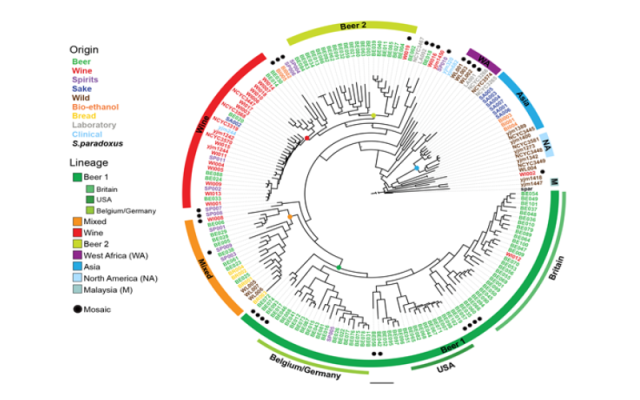
We investigate how genetic and phenotypic differences arise and how they are affected by evolutionary processes such as domestication and hybridization. We have a collection of more than 600 sequenced yeasts isolated from multiple industrial (beer, wine, bioethanol etc.) and natural environments and we routinely isolate and sequence new yeasts. In addition, we use high-throughput phenotyping to measure hundreds of traits.
Ultimately, our goal is to identify, to single-nucleotide resolution, the genetic variation associated to industrially relevant traits such as flavor production and ethanol tolerance.